SyCLoPS User Manual¶
The SyCLoPS paper: The System for Classification of Low-Pressure Systems (SyCLoPS): An All-in-One Objective Framework for Large-scale Datasets (Han & Ullrich, 2025). Link to the article: https://agupubs.onlinelibrary.wiley.com/doi/10.1029/2024JD041287
Recent updates in SyCLoPS are documented in this preprint: Link to the article
Documentation about the TempestExtremes(TE) software can be found HERE. Missing values in datasets can be safely handled by TE in the latest realease.
All the SyCLoPS codes and comments can be found in on SyCLoPS @ GitHub.
Published Data:
SyCLoPS ERA5 LPS Catalogs @ Zenodo
SyCLoPS HighResMIP LPS Catalogs (No TLCs) @ Zenodo
SyCLoPS HighResMIP LPS Catalogs (TC-only) @ Zenodo
Table of Contents:
- Look-up Tables and the Classification Flowchart (1.1-1.6)
- Tips for Installing TE (2.1)
- Tips for using SyCLoPS TE command lines (2.2)
- Tips for using SyCLoPS with different climate and model data (2.3)
- Tips for using SyCLoPS with regional model outputs (2.4)
- Tips on tracking LPSs on unstructured grids (2.5)
- SyCLoPS Catalogs Usages and Applications (3.1-3.2)
The two main steps to implement SyCLoPS are:
Review the codes and comments in
SyCLoPS_main.pyon GitHub. Change variable names and other specifications according to your needs.Run
SyCLoPS_main.pyand follow the instructions carefully.
There are several optional steps for SyCLoPS applications which are discussed in section 3.2 of this manual.
If this manual does not answer your questions about SyCLoPS, please contact the author Yushan Han at yshhan@ucdavis.edu
1. Look-up Tables and the Classification Flowchart¶
This section reproduces several tables and the SyCLoPS workflow diagram from the SyCLoPS manuscript with some more details.
1.1 Variable requirements¶
| Variable Name | Pressure Level (hPa) |
|---|---|
| U-component Wind (U) | 925, 850, 200$^{a}$, 500 |
| V-component Wind (V) | 925, 850, 200$^{a}$, 500 |
| Temperature (T) | 850 |
| Relative Humidity (R)$^{b}$ | 850, 100$^{c}$ |
| Mean Sea Level Pressure (MSL) | Sea Level |
| Geopotential (Z) / Height (H) | Surface (invariant)$^{d}$, 925, 850$^{d}$, 700, 500, 300$^{a}$ |
a. These default levels of U,V,Z can be replaced with the more commonly found 250-hPa level. See Appendix B in the SyCLoPS paper for details.
b. Specific humidity can be converted to relative humidity (R) with temperature data at 850 and 100 hPa. To get the R at 100 hPa accurately, you may need to convert it as the relative humidity with respect to ice.
c. The daily frequency for R at 100 hPa is sufficient to maintain good performance of the SyCLoPS classification. See Appendix B in the SyCLoPS paper for details.
d. Z or H at 850 hPa and the surface level is optional if the data set has missing/fill values where the data plane intersects the surface. See comments in "SyCLoPS_main.py."
P.S. 10 m U and V component wind variables (VAR_10U,VAR_10V) are optional and can be used to calculate the maximum surface wind speed (WS) of an Low-Pressure System (LPS) as reference information in the classified catalog. They do not affect the detection and classification process.
1.2 LPS Initialism Table¶
Initialism |
Full Term |
Definition |
|---|---|---|
| HAL | High-altitude Low | LPSs found at high altitudes without a warm core |
| THL | Thermal low | Shallow systems featuring a dry and warm lower core |
| HATHL | High-altitude Thermal Low | LPSs found at high altitudes with a warm core |
| DOTHL | Deep (Orographic) Thermal Low | Non-shallow LPSs featuring a dry and warm lower core often driven by topography |
| TC | Tropical Cyclone | LPSs that would be named in IBTrACS |
| TD | Tropical Depression | Tropical systems that have developed a weak upper-level warm core and are strong enough to be recorded as TDs in IBTrACS |
| TLO | Tropical Low | Non-shallow tropical systems that fall short of TD requirements |
| MD | Monsoon Depression | TDs developing in monsoonal environment. A monsoon environment is considered to be dominated by westerly winds (resulting in asymmetric wind fields in monsoon LPS) and very humid Labeled as "TD(MD)" in the classified catalog. TDs that fall short of the monsoonal system condition are labeled "TD" |
| ML | Monsoon Low | TLOs developing in monsoonal environment. Labeled as "TLO(ML)" in the classified catalog. TLOs that fall short of the monsoonal system condition are labeled "TLO" |
| MS | Monsoonal System | Monsoon LPSs (MDs plus MLs) |
| TLC | Tropical-Like Cyclone | Non-tropical LPSs that resemble TCs (typically smaller than TCs). For example, they can have gale-force sustained surface wind, well-organized convection (sometimes with an eyewall) and a deep warm core |
| SS (STLC) | Subtropical Storm (Subtropical Tropical-Like Cyclone) | A type of TLC in the subtropics, represented by Mediterranean hurricanes |
| PL (PTLC) | Polar Low (Polar Tropical-Like Cyclone) | A type of TLC typically found north of the polar front |
| SC | Subtropical Cyclone | A type of LPS that is typically associated with a upper-level cut-off low south of the polar jet and has a shallow warm core |
| EX | Extratropical Cyclone | Most typical non-tropical cyclones |
| DS | Disturbance | Shallow LPSs or waves with weak surface circulations. DSD, DST and DSE are dry, tropical and extratropical DSs |
| QS | Quasi-stationary | LPSs that stay relatively localized as labeled by the QS track condition |
1.3 The Input LPS Catalog Column Table (for SyCLoPS_input.parquet)¶
| Column | Unit |
Description |
|---|---|---|
| TID | - | LPS track ID (0-based) in both the input and classified catalog |
| ISOTIME | - | UTC timestamp (ISO time) of the LPS node in both catalogs |
| LON | ° | Longitude of the LPS node in both catalogs |
| LAT | ° | Latitude of the LPS node in both catalogs |
| MSLP | Pa | Mean sea level pressure at the LPS node in both catalogs |
| MSLPCC20 | Pa | Greatest positive closed contour delta of MSLP over a 2.0° Great Circle Distance (GCD), representing the core region of an LPS |
| MSLPCC55 | Pa | Greatest positive closed contour delta of MSLP over a 5.5° GCD |
| DEEPSHEAR | $\mathrm{m\:s^{-1}}$ | Average deep-layer wind speed shear between 200 hPa and 850 hPa over a 10.0° GCD |
| UPPTKCC | $\mathrm{m^{2}\:s^{-2}}$ | Greatest negative closed contour delta of the upper-level thickness between 300 hPa and 500 hPa over a 6.5° GCD, referenced to the maximum value within 1.0° GCD |
| MIDTKCC | $\mathrm{m^{2}\:s^{-2}}$ | Greatest negative closed contour delta of the middle-level thickness between 500 hPa and 700 hPa over a 3.5° GCD, referenced to the maximum value within 1.0° GCD |
| LOWTKCC$^{a}$ | $\mathrm{m^{2}\:s^{-2}}$ | Greatest negative closed contour delta of the lower-level thickness between 700 hPa and 925 hPa over a 3.5° GCD, referenced to the maximum value within 1.0° GCD |
| Z500CC | $\mathrm{m^2\:s^{-2}}$ | Greatest positive closed contour delta of geopotential at 500 hPa over a 3.5° GCD referenced to the minimum value within 1.0° GCD |
| VO500AVG | $\mathrm{s^{-1}}$ | Average relative vorticity over a 2.5° GCD |
| RH100MAX | % | Maximum relative humidity at 100 hPa within 2.5° GCD |
| RH850AVG | % | Average relative humidity over a 2.5° GCD at 850 hPa |
| T850 | K | Air temperature at 850 hPa at the LPS node |
| Z850 | $\mathrm{m^2\:s^{-2}}$ | Geopotential at 850 hPa at the LPS node |
| ZS | $\mathrm{m^2\:s^{-2}}$ | Geopotential at the surface at the LPS node |
| U850DIFF | $\mathrm{m\:s^{-1}\:sr}$ | Difference between the weighted area mean of positive and negative values of 850 hPa U-component wind over a 5.5° GCD |
| WS200PMX | $\mathrm{m\:s^{-1}}$ | Maximum poleward value of 200 hPa wind speed within 1.0° GCD longitude |
| RAWAREA$^{b}$ | $\mathrm{km^2}$ | The raw defined size (see appendix E) of the LPS |
| LPSAREA | $\mathrm{km^2}$ | The adjusted defined size of the LPS in both catalogs (see appendix E) |
a. 925 hPa may be replaced by 850 hPa if data at this level are not available in some datasets.
b. This parameter in the column is for user reference only. It does not affect any results in the SyCLoPS LPS node or track classification.
1.4 Classification Condition Table¶
| Condition Name | Conditions |
|---|---|
| High-altitude Condition$^{a}$ | Z850 > ZS |
| Dryness Condition | RH850AVG < 60% |
| Cyclonic Condition | VO500AVG >= (<) 0 $\mathrm{s^{-1}}$ if LAT >= (<) 0° |
| Tropical Condition | RH100MAX > 20%; DEEPSHEAR < 18 $\mathrm{m\:s^{-1}}$; T850 > 280 K |
| Transition Condition | Tropical Conditon = True; DEEPSHEAR $>$ 10 $\mathrm{m\:s^{-1}}$ or RH100MAX < 55% |
| TC Condition$^{b}$ | MSLPCC20 > 215 Pa (Variable); LOWTKCC < 0 $\mathrm{m^2\:s^{-2}}$; UPPTKCC < -107.8 $\mathrm{m^2\:s^{-2}}$ |
| TD Condition | MSLPCC55 > 160 Pa; UPPTKCC < 0 $\mathrm{m^2\:s^{-2}}$ |
| MS Condition | U850DIFF > 0 $\mathrm{m\:s^{-1}}$; RH850AVG > 85% |
| TLC Condition$^{c}$ | MSLPCC20 > 190 Pa; LOWTKCC and MIDTKCC $<$ 0 $\mathrm{m^2\:s^{-2}}$;(LPSAREA < 5.5 × 105 km^2; LPSAREA > 0 km^2) or (MSLPCC20 > 420 Pa; MSLPCC20 : MSLPCC55 > 0.5) |
| SC Condition | LOWTKCC < 0 $\mathrm{m^2\:s^{-2}}$; Z500CC $>$ 0 $\mathrm{m^2\:s^{-2}}$;WS200PMXc > 30 $\mathrm{m\:s^{-1}}$ |
| TC Track Condition | At least 8 (8+) 3-hourly TC-labeled nodes in an LPS track |
| MS Track Condition | 10+ 3-hourly "TLO(ML)" or "TD(MD)"-labeled nodes |
| SS Track Condition | 2+ 3-hourly TLC-labeled nodes ("SS(STLC)" or "PL(PTLC)") |
| PL Track Condition | 2+ 3-hourly TLC-labeled nodes ("SS(STLC)" or "PL(PTLC)") |
| QS Track Condition | See SI text S3 for details |
a. It can be as simple as checking the availability of T850 (or Z850) data (have null/missing values or not) in some records.
b. TC condition is optimized automatically by both model grid resolution and the pressure level used (e.g. 250/200 hPa).
c. See section 5.3 in the SyCLoPS paper for a possible alternative.
1.5 The Classified LPS Catalog Column Table (for SyCLoPS_classified.parquet)¶
| Column | Unit |
Description |
|---|---|---|
| TID | - | LPS track ID (0-based) in both the input and classified catalog |
| ISOTIME | - | UTC timestamp (ISO time) of the LPS node in both catalogs |
| LON | ° | Longitude of the LPS node in both catalogs |
| LAT | ° | Latitude of the LPS node in both catalogs |
| MSLP | Pa | Mean sea level pressure at the LPS node in both catalogs |
| WS* | $\mathrm{m\:s^{-1}}$ | Maximum wind speed at the 10-m level within 2.0° GCD |
| Full_Name | - | The full LPS name based on the classification |
| Short_Label | - | The assigned LPS label (the abbreviation of the full name) |
| Tropical_Flag | - | 1 if the LPS is designated as a tropical system, otherwise 0 |
| Transition_Zone | - | 1 if the LPS is in the defined transition zone, otherwise 0 |
| Track_Info | - | "TC", "MS", "SS(STLC)", "PL(PTLC)", "QS" denoted for TC, MS, SS, PL and QS tracks; "EXT", "TT" denoted for extratropical and tropical transition completion nodes |
| IKE* | $\mathrm{TJ}$ | The integrated kinetic energy computed based on the LPS size blobs that are used to define RAWAREA |
* These two columns are for user reference only. They do not affect any results in the SyCLoPS LPS node or track classification.
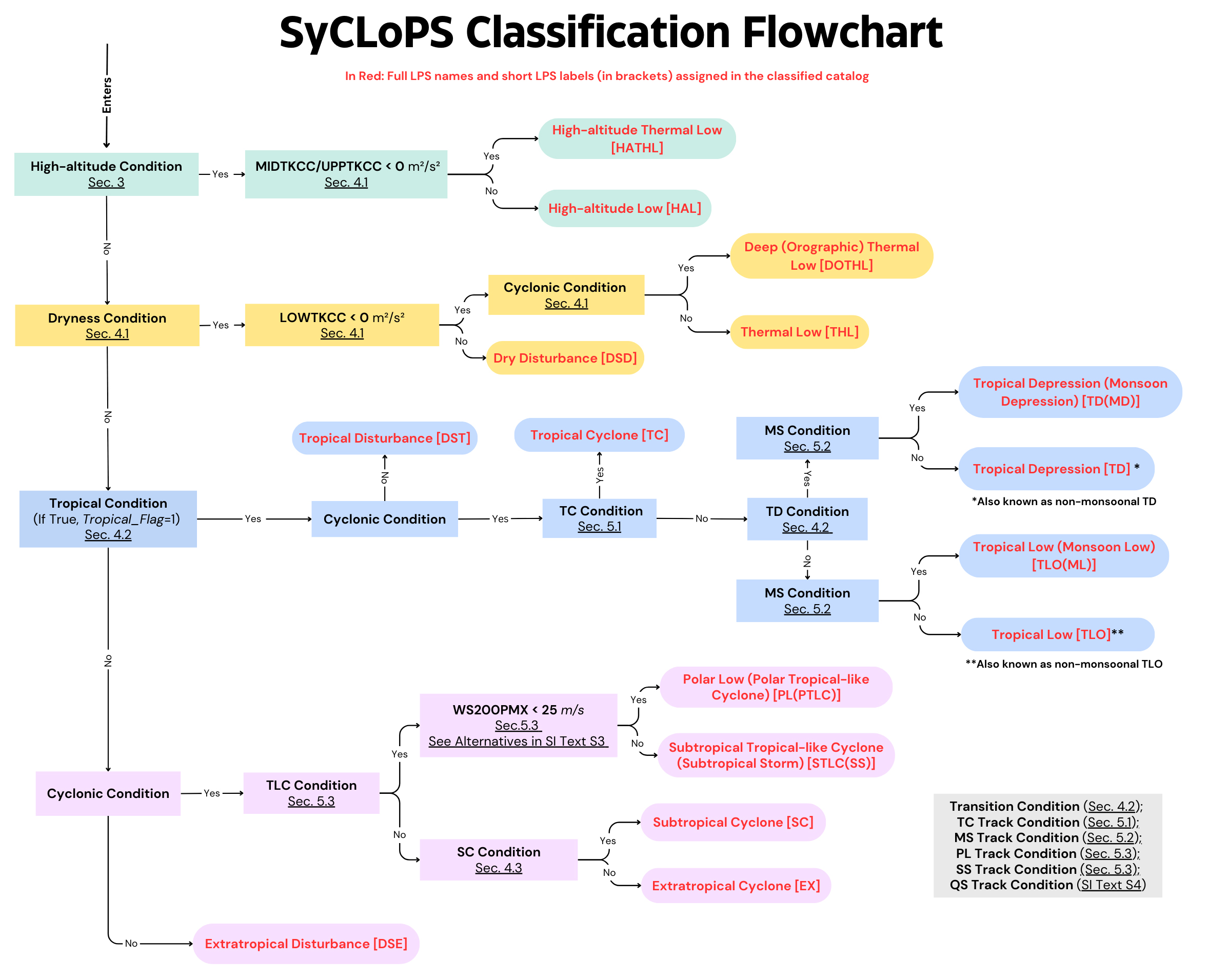
1.6 SyCLoPS Classification Flowchart and Assigned Labels and Full Names¶
Section numbers in the figure refer to the section numbers in the SyCLoPS manuscript. From top to bottom, there are four branches: High-altitude, dry, tropical, and extratropical branches.
2. Tips for running SyCLoPS¶
2.1 Installing TE¶
TE is loacted at: https://github.com/ClimateGlobalChange/tempestextremes. Missing values in datasets can be safely handled by TE in the latest realease.
It is recommended that you download/clone and compile TE using the provided
quick_make_general.shfile, as TE may not be updated frequently on the conda channel and it may have problems with MPI-enabled installtion.Please refer to the TE documentation for detailed explanations of each function, argument, and operation: https://climate.ucdavis.edu/tempestextremes.php
To install and run TE in parallel, please make sure that your computer has an MPI implementation installed (e.g., Open MPI). When using MPI in TE commands, simply add
srun -n 128ormpirun -np 128to the beginning of some TE commands to enable parallel computation. For more details, see the MPI documentation provided online or by your supercomputer host.DetectNodes, DetectBlobs and VariableProcessor support MPI computation in TE version 2.3.x. The parallelization is achieved by writing the inputfile as a list of files, each containing variables of a time slice. See the next subsection for details.
If you have any questions about installing the TE software, please contact Paul Ullrich at paullrich@ucdavis.edu.
2.2 TE Operations explained in the SyCLoPS main program¶
You will first need to specify the installation path of TE on your computer and adjust other specifications in the beginning of the srcipt (
SyCLoPS_main.py) mannually, if necessary.Prepare a list of files (in txt format) containing all the required variables (see table in 1.1) required by the program as the inputfile. The files in the list should be arranged in time slices, like this:
Variable1_TimeSlice1;Variable2_TimeSlice1;Variable3_TimeSlice1;...
Variable1_TimeSlice2;Variable2_time_slice2;Variable3_TimeSlice2;...
Variable1_TimeSlice3;Variable2_time_slice3;Variable3_TimeSlice3;...
...
It is recommended that you use MSLP files as your "Variable 1" at the beginning of each row in your input file list to set the time resolution of your detection. This is because TE uses the time series of the first file in a row of a list to determine the time resolution of the detection. Please also make sure that all the following files in each row contain the time steps present in the first file. That means, your "Variable 2" and beyond can have longer time periods than "Variable 1" as long as they contain the required time steps set by Variable 1.The output files (outputfile) used throughout the program should contain the same number of lines as the inputfile. So it should have a filename for each time slice on each line, which should look like this:
ERA5_LPSnode_out_TimeSlice1
ERA5_LPSnode_out_TimeSlice2
ERA5_LPSnode_out_TimeSlice3
...Below is a sample shell script to list 4 different variables (MSL, Z, U and ZS (the constant surface geopotential)) with different time slices to generate the input file along with a corresponding output file (the outputfile is generated simultaneously):
#!/bin/bash
ERA5DIR=/global/cfs/projectdirs/m3522/cmip6/ERA5
mkdir -p LPS
rm -rf ERA5_example_in.txt # the input file
rm -rf ERA5_example_out.txt # the output file
for f in $ERA5DIR/e5.oper.an.pl/*; do
# In this example ERA5 directory, variables are stored in folders named by years and months (e.g., 202001,202002)
yearmonth=$(basename $f)
year=${yearmonth:0:4}
echo "..${yearmonth}"
if [[ $year -gt '1978' ]] && [[ $year -lt '2023' ]]
then
for zfile in $f/*128_129_z*; do
zfilebase=$(basename $zfile)
yearmonthday=${zfilebase:32:8}
mslfile=`ls $ERA5DIR/e5.oper.an.sfc/${yearmonth}/*128_151_msl*`
ufile=`ls $ERA5DIR/e5.oper.an.pl/${yearmonth}/*128_131_u.*${yearmonthday}*`
zsfile=./e5.oper.invariant.Zs.ll025sc.nc
echo "$mslfile;$zfile;$ufile;$zsfile" >> ERA5_example_in.txt
echo "LPS/era5.LPS.node.${yearmonthday}.txt" >> ERA5_example_out.txt
done
fi
done
- Here's another example shell script to list 4 different variables (Z, MSL, U10 and ZS, the constant surface geopotential) with different time slices for the inputfile in a customized data directory with filenames containing data's time period (e.g., 20100101):
#!/bin/bash
DIR="/path/to/your/folder"
# Extract unique dates from filenames
dates=$(ls "$DIR" | grep -oP '\d{8}' | sort -u)
for date in $dates; do
echo "msl_${date}.nc;u10_${date}.nc;z_${date}.nc;zs_${date}"
done
If you use MPI in applicable TE commands, each thread will take one time slice (row) in the
$inputfileat a time and output a corresponding output file. You will need to setuse_srun = TrueinSyCLoPS_main.py.If you use variables on a unstructured grid (i.e., not lon-lat grid), you will need to set
use_connect = Trueand specify a connectivity file inSyCLoPS_main.py. Some more detalis on how to generate a TE-compatible connectivty file can be found in section 2.5 below.If you use 250 hPa data instead of the default 200/300 hPa fields, set
use_250hPa_only = True. See section 2.3 below for details.DetectNodes: This command detects candidate LPS nodes and computes the 15 parameters needed for the classification:
This step can be time consuming. It's highly recommended to run this command in parallel, feeding it with a list of files ordered by time slices (see section 2.2). "ZS" is not needed if your data has missing values where the 850 hPa data level is below the surface.
The time dimension in the invariant surface geopotential file for ZS/Z0 should be removed (averaged) prior to the following procedures (if they have not already). It can be achieved by something like: "ncwa -a time ZS_in.nc ZS_out.nc."
"WS" (the near-surface maximum wind speed within 2.0 GCD of the LPS node) is an optional parameter for reference purporse. Add
_VECMAG(VAR_10U,VAR_10V),max,2.0to the end of the--outputcmdif you want to output this parameter. "Z850" is also not needed if your data has missing values where the 850 hPa data level is below the surface.If you want to focus on life cycles of tropical-like cyclones (Polar Low (PL) and Subtropical Storm (SS)): Due to their small sizes, it's recommended to lower the merging distance (
MergeDist) from 6.0 GCD to ~3.0 GCD.
StitchNodes: This command stitches all detected nodes in sequence with parameters formatted in a csv file.
If you specify a range distance (
RangeDistfor the--rangeargument) that is larger than the Merging distance that you have set for DetectNodes, the software will automatically adds the new--prioritize MSLPargument to the end. The--prioritizeargument prevents potential false connections at the supposed end of a track in this scenario. See more details in the supporting information text S2 in our preprint.If you are using a 6-hour detection rate in DetectNodes, you should consider either doubling the "4.0" in the
RangeDist(used for the default 3-hourly resolution) to "8.0" or just using "6.0." A 6.0 GCD range is considered sufficient to cover tropical systems, but it might be insufficient to track some extremely fast-moving extratropical cyclones. A range distance of 8.0 GCD produces longer track length, but it might contain a few false connections connecting the supposed end of a track to another system's track. Therefore, it's recommended to use "6.0" if you only care about the tropical phase of tropical system tracks.If you are using a 6-hour detection rate in DetectNodes, you should also consider lowering the
MSLPCC55CCStepfrom 5 to 3 or 2.if you are using shorter detection frequency (e.g., 1-hourly), please adjust the above parameters at your discretion.
You may also adjust other parameters in DetectNodes and StitchNodes to meet your end needs.
TE commands 9-11 are reserved for computing LPSAREA and generating blob files. They can be omitted if you are not labeling tropical-like cyclones (TLCs) and precipitation/wind blobs. You may choose to skip these steps when prompted in the main program. You can also choose to skip the steps related to computing LPSAREA and labeling TLCs (SSs and PLs) in SyCLoPS_classifier.py when prompted. In this case, the program will only assign SC/EX/DSE labels if the LPS node is classified in the extratropical branch.
VariableProcessors: It calcualtes the cyclonic relative vorticity by computing a smoothed 850 hPa relative vorticity (RV) field and revert the sign of RV in the southern Hemisphere to get the cyclonic RV. This command can be run in parallel. Your inputfile should conatin files of U and V at 850 hPa and 925 hPa. If you are specifically interested in LPSs close to mountainous regions above 850 hPa level, it's recommended that you use 700 hPa data instead.
DetectBlobs: This step is to generate LPS size blobs. It's recommended to run this command in parallel, feeding it with a list of files ordered by time slices. It's possible to use 850 and 700 hPa wind speed to replace the 925 hPa, and it's recommended to slightly increase the wind speed threshold used in this step in this scenario. A similar tactic to generate LPS precipitation blobs is introdcued in the
TE_optional.shfile on Zenodo.BlobStats: Generate useful information for calculating LPSAREA and tagging blobs with labels. This step cannot be run in parallel but could potentially be time-consuming if you use "sumvar" to compute IKE of each LPS for reference purposes (note that IKE is not required for classification). In this case, one may opt to use the GNU parallel tool to run multiple commands simultaneously, each using a single thread. To accomplish this, users should first create a txt file containing a list of TE commands to be parallelized (e.g. breaking down into years) and a list of corresponding input and output files. Then run
parallel -j n < blobstats_commands_list.txt("n" is the number of threads to use) to start the paralled BlobStats processes aftermodule load parallel. You would also need to add files of U and V at 925 hPa to the inputfile list for IKE computation.The Classifier: After running all TE commands, the program will prompt you to procceed with executing
SyCLoPS_classifier.pywhen you are ready. You would first need to check the comments inSyCLoPS_classifier.pyand change the specifications at the beginning of the classifier script if necessary. Note: You would also need to enter the model/data's grid resolution in deg^2 at the beginning of the script to achieve optimized TC detection skills.
2.3 Tips on applying SyCLoPS to climate and model data¶
Most high-resolution climate model outputs do not have all the required variables at 3 hours resolution, but they are mostly available at 6-hourly resolution, which is good enough for tracking most LPSs. However, relative humidity (RH) at 100 hPa is usally only available at a daily-mean basis. In the SyCLoPS paper, we show that using daily-mean relative humidity or 6-hourly detection rate will not lower SyCLoPS performance (except that TLC detection skill decreases as detection rate decreases). You can now directly use daily RH files together with files in other time resolutions in TE's calculations (the latest TE release needed). No oversampling needed.
Climate model outputs may not contain the 300 hPa and 200 hPa data used in the default settings of SyCLoPS, but they typically have 250 hPa data available. We have also shown in the paper that using 250 hPa data instead of 200 hPa and 300 hPa does not degrade SyCLoPS performance with some minor adjustments (see Appendix B of the paper). To use 250 hPa data to replace 200/300 hPa data, specify
use_250hPa_only = TrueinSyCLoPS_main.pyand type "Y(Yes)" when asked if you want to use 250 hPa data instead of the default 200/300 hPa when runningSyCLoPS_classifier.py.
2.4 Tips on applying SyCLoPS to regional model data¶
Because of the nature of the closed contour critera used in SyCLoPS and TE, false LPS tracks will be detected near the four edges of the regional domain. Hence, it is recommended to define a ~2° buffer zone sourrounding the four boundaries of your regional domain to remove tracks in that zone in a post-processing process.
Another option is to define
--minlat,--maxlat,--minlon,--maxlonin the DetectNodes command. If your domain boundaries are not parallel to latitude or longitude, you can create a mask file as an input to DetectNodes to define a detection zone of your domain (with the grid labeled "1" in that zone). Then add something likeZONE_MASK,=,1,0.0to--thresholdcmdin the DetectNodes command.When running
SyCLoPS_classifier.py, type "Y(Yes)" when asked if you are running with regional model data, so that the program will opt to use the alternative criteria designed for regional models (see SyCLoPS Supporting Information S4 for details) at several points in the classification process.Some models may only output specific humidity. In this case, you can use specific humidity and temperature to calculate relative humidity. Remember to calculate RH with respect to ice at 100 hPa.
2.5 Tips on tracking LPSs on unstructured grids¶
A SCRIP format mesh NetCDF file is required for TE to generate the connectivity file used for tracking LPSs seamlessly on unstructured grids. This mesh file is often provided by the model producer. Please see an example of MPAS models below:
- Choose the desired MPAS mesh file from here. For variable mesh grids, you will use the file with your refinement region after grid rotation (see MPAS-A Users' Guide for instructions).
- Because the MPAS C-grid mesh file is in its own format rather than the SCRIP format we want. It needs to be converted using the MPAS-tool package. You would first need to install mpas_tool via conda-forge:
conda create -n mpas_tools_env -c conda-forge mpas_tools, and then doconda activate mpas_tools_env. - Use the built-in tool scrip_from_mpas to convert files:
scrip_from_mpas -m MPAS_Filename -s Output_SCRIP_Filename
Once you have the mesh file, you can run the following TE command lines to generate the connectivity file:
#!/bin/bash
TEMPESTEXTREMES_DIR=/path/to/your/TempestExtremes/directory
SCRIP_Mesh_file=/path/to/your/mesh_file
Output_connectivity_file=/path/to/your/output_connectivity_file
$TEMPESTEXTREMES_DIR/GenerateConnectivityFile \
--in_mesh ${SCRIP_Mesh_file} \
--out_connect ${Output_connectivity_file} \
- You can now use the connectivity file as directed above in Point 5 of section 2.2.
3. SyCLoPS Catalogs Usages and Applications¶
3.1 How to select different types of LPS nodes and tracks in the classified catalog:¶
Please see Task 3 and Task 6 for selecting all tropical cyclone (TC) tracks/nodes and all extratropical (non-tropical) cyclone nodes, respectively.
We recommend using the Adjusted_Label column for the final LPS label references. This column is adjusted for short-term, unstable classifications in boundary cases of the tropical cyclone (TC) and monsoonal system (MS) tracks. This adjustment improves classification accuracy (See our preprint's supporting information text S3 for details). Due to this adjustment, you may occasionally find that the labels in the Adjusted_Label column differ from those in the Short_Label column. The Full_Name column is not provided anymore. Please refer to the initialism table in section 1.2 for full names of the LPS labels.
To open the classified catalog:
import numpy as np
import pandas as pd
ClassifiedCata='SyCLoPS_classified_ERA5_1940_2024.parquet' # your path to the classified catalog
dfc=pd.read_parquet(ClassifiedCata) # open the parquet format file. PyArrow package required.
model_data_name = 'ERA5_1940_2024' # your model data name, e.g., ERA5_1940_2024
inputfile_DN = f"file_list/{model_data_name}_lpsnode_in.txt"
inputfile_DN2 = inputfile_DN
dfc
| TID | LON | LAT | ISOTIME | MSLP | WS | ZS | Short_Label | Adjusted_Label | Tropical_Flag | Transition_Zone | Track_Info | LPSAREA | i | j | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 38.00 | 74.75 | 1940-01-01 00:00:00 | 99764.19 | 18.17691 | -0.813965 | EX | EX | 0.0 | 0.0 | Track | 0 | 152 | 61 |
| 1 | 0 | 38.75 | 75.25 | 1940-01-01 03:00:00 | 99729.75 | 19.37490 | 3.150879 | EX | EX | 0.0 | 0.0 | Track | 0 | 155 | 59 |
| 2 | 0 | 37.00 | 75.50 | 1940-01-01 06:00:00 | 99653.75 | 19.31102 | -8.552246 | EX | EX | 0.0 | 0.0 | Track | 0 | 148 | 58 |
| 3 | 0 | 35.00 | 75.50 | 1940-01-01 09:00:00 | 99589.88 | 19.72554 | 5.807129 | EX | EX | 0.0 | 0.0 | Track | 0 | 140 | 58 |
| 4 | 0 | 33.25 | 75.25 | 1940-01-01 12:00:00 | 99357.12 | 19.69311 | 6.877441 | EX | EX | 0.0 | 0.0 | Track | 0 | 133 | 59 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 15048189 | 740026 | 257.50 | -71.50 | 2024-12-31 09:00:00 | 95383.69 | 19.85738 | -20.356930 | EX | EX | 0.0 | 0.0 | Track | 1461492 | 1030 | 646 |
| 15048190 | 740026 | 256.75 | -71.75 | 2024-12-31 12:00:00 | 95496.31 | 17.94809 | -44.622560 | EX | EX | 0.0 | 0.0 | Track | 1379330 | 1027 | 647 |
| 15048191 | 740026 | 252.50 | -72.75 | 2024-12-31 15:00:00 | 95605.81 | 16.39059 | 2.424316 | EX | EX | 0.0 | 0.0 | Track | 1282608 | 1010 | 651 |
| 15048192 | 740026 | 251.25 | -73.00 | 2024-12-31 18:00:00 | 95773.50 | 15.88286 | -5.603027 | EX | EX | 0.0 | 0.0 | Track | 1154036 | 1005 | 652 |
| 15048193 | 740026 | 250.25 | -73.25 | 2024-12-31 21:00:00 | 95974.94 | 14.95377 | -1.739746 | EX | EX | 0.0 | 0.0 | Track | 973231 | 1001 | 653 |
15048194 rows × 15 columns
If desired, the input and ouput (classified) catalogs can also be combined to produce a larger catalog:
# InputCata='SyCLoPS_input.parquet'
# dfin=pd.read_parquet(InputCata)
# dfc=pd.concat([dfc,dfin],axis=1)
Task 1. Select a single type of LPS node (e.g., Thermal Low):
dftc=dfc[dfc.Adusted_Label=='THL']
Task 2. Select two types of LPS node (e.g., EX and SC):
dfexsc=dfc[(dfc.Adjusted_Label=='EX') | (dfc.Adjusted_Label=='SC')]
Task 3. Select all TC tracks and all TC nodes (the reocmmended way):
# TC Tracks:
dftc_track=dfc[dfc.Track_Info.str.contains('TC')]
# Print TC track ids:
tctid=pd.unique(dftc.TID)
print(tctid)
# TC Nodes:
dftc_node=dfc[dfc.Track_Info.str.contains('TC') & dfc.Adjusted_Label=='TC']
Task 4. Select Monsoonal system nodes in MS tracks:
dfms_node=dfc[dfc.Track_Info.str.contains('M') & dfc.Adjusted_Label.str.contains('M')]
Task 5. Define and select the TC stage of the tracks (the first TC node to the last TC node of each track):
tc_sections = []
for tid, group in dftc.groupby('TID'):
tc_indices = group.index[group.Short_Label == 'TC']
start, end = tc_indices[0], tc_indices[-1]
section = dfc.loc[start:end]
tc_sections.append(section)
dftc_tcstage = pd.concat(tc_sections)
# Similarly you can define a pre-TC and post-TC stage of the TC tracks.
Task 6. Select extratropical (non-tropical) cyclones of all kinds:
dfexnode=dfc[(dfc.Tropical_Flag==0)]
Task 7. Select two types of TLC node (including SS(STLC) and PL(PTLC)):
dftlc=dfc[dfc.Adjusted_Label.str.contains('TLC')]
dftlc
| TID | LON | LAT | ISOTIME | MSLP | WS | ZS | Short_Label | Adjusted_Label | Tropical_Flag | Transition_Zone | Track_Info | LPSAREA | i | j | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 45 | 1 | 40.00 | 53.00 | 1940-01-05 12:00:00 | 100427.60 | 7.732711 | 1411.874000 | PL(PTLC) | PL(PTLC) | 0.0 | 0.0 | Track | 468842 | 160 | 148 |
| 98 | 6 | 56.00 | 58.50 | 1940-01-02 06:00:00 | 100405.90 | 6.140673 | 1592.909000 | PL(PTLC) | PL(PTLC) | 0.0 | 0.0 | Track_PL(PTLC) | 333618 | 224 | 126 |
| 99 | 6 | 56.00 | 58.75 | 1940-01-02 09:00:00 | 100456.40 | 6.009427 | 1362.959000 | PL(PTLC) | PL(PTLC) | 0.0 | 0.0 | Track_PL(PTLC) | 172463 | 224 | 125 |
| 170 | 11 | 207.75 | 58.50 | 1940-01-03 12:00:00 | 99349.31 | 13.449820 | 545.260300 | SS(STLC) | SS(STLC) | 0.0 | 0.0 | Track | 23202 | 831 | 126 |
| 216 | 14 | 161.75 | 50.75 | 1940-01-02 12:00:00 | 98858.94 | 20.418770 | 1.150879 | SS(STLC) | SS(STLC) | 0.0 | 0.0 | Track | 0 | 647 | 157 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 15048112 | 740017 | 188.25 | 41.75 | 2024-12-31 09:00:00 | 98137.94 | 26.965960 | 1.881348 | SS(STLC) | SS(STLC) | 0.0 | 0.0 | Track_SS(STLC)_PL(PTLC) | 1591329 | 753 | 193 |
| 15048113 | 740017 | 189.75 | 42.75 | 2024-12-31 12:00:00 | 98199.81 | 24.381820 | 0.271973 | SS(STLC) | SS(STLC) | 0.0 | 0.0 | Track_SS(STLC)_PL(PTLC) | 1734546 | 759 | 189 |
| 15048114 | 740017 | 191.00 | 43.50 | 2024-12-31 15:00:00 | 98066.06 | 23.533540 | -0.981934 | PL(PTLC) | PL(PTLC) | 0.0 | 0.0 | Track_SS(STLC)_PL(PTLC) | 1886487 | 764 | 186 |
| 15048115 | 740017 | 191.75 | 44.00 | 2024-12-31 18:00:00 | 98096.75 | 23.149840 | 0.549316 | PL(PTLC) | PL(PTLC) | 0.0 | 0.0 | Track_SS(STLC)_PL(PTLC) | 2204847 | 767 | 184 |
| 15048116 | 740017 | 192.00 | 44.25 | 2024-12-31 21:00:00 | 98300.44 | 23.402600 | -3.817871 | PL(PTLC) | PL(PTLC) | 0.0 | 0.0 | Track_SS(STLC)_PL(PTLC) | 2163477 | 768 | 183 |
409594 rows × 15 columns
Task 8. Select all DST and all TLO (TLO and TLO(ML)) nodes in TC tracks:
dftc3=dfc[((dfc.Adjusted_Label=='DST')|(dfc.Adjusted_Label.str.contains('TLO'))) & (dfc.Track_Info.str.contains('TC'))]
Task 9. Select LPS track IDs (TIDs) that have at least 5 non-tropical LPS nodes that are not DSE:
dfex=dfc[(dfc.Tropical_Flag==0)&(dfc.Adjusted_Label!='DSE')]
extrackid=pd.unique(dfex.TID)[dfex.groupby('TID')['TID'].count()>=5]
Task 10. Select Tracks that are both a TC track, SS track and PL(PTLC) track:
tcsspl_trackid=pd.unique(dfc[(dfc.Track_Info.str.contains('TC')) & (dfc.Track_Info.str.contains('SS')) & (dfc.Track_Info.str.contains('PL'))].TID)
dftcsspl=dfc[dfc.TID.isin(tcsspl_trackid)]
Task 11. Select all tropical TCs nodes in TC tracks that are not undergoing extratropical transition:
dftc3=dfc[(dfc.Track_Info.str.contains('TC')) & (dfc.Tropical_Flag==1) & (dfc.Transition_Zone==0)]
Task 12. Select all tropical transition completion nodes:
dftt=dfc[dfc.Track_Info.str.contains('TT')]
Task 13. Select TC tracks that do not undergo extratropical transition:
tcnoext_trackid=pd.unique(dfc[(dfc.Track_Info.str.contains('TC')) & ~(dfc.Track_Info.str.contains('EXT'))].TID)
Task 14. Select all LPS nodes within a bounded region in January:
dflps=dfc[(dfc.LAT>=30) & (dfc.LAT<=50) & (dfc.LON>=280) & (dfc.LON<=350) & (dfc.ISOTIME.dt.month==1)]
3.2 Other applications based on the classified catalog:¶
Here we introduce two additional usages of SyCLoPS: calculating intergataed kinetic energy (IKE) accumulation or precipitation contribution percentage for a specific type of LPS.
To perform this task, users need to run Blob_idtag.py and TE_optional.sh. Users can opt to run the additional TE commands within `Blob_idtag_app.py' (lines 13-15). The procedure can be divided into five steps:
- The additional TE commands in
TE_optional.shdetect precipitation blobs and calculate blob statistics (properties) using BlobStats in addition to the size blobs already detected inSyCLoPS_main.py. - Both size and precipitation blobs are masked with a unique ID (1-based, e.g., 1,,2,3,4,5,...) through StitchBlobs.
- The blob-tagging Python script (
Blob_idtag.py) pairs precipitation blobs to LPS nodes in the same way as we did for size blobs. - The Python script assigns tags (labels) to different blobs according to their paired labeled LPS nodes and the blobs IDs given by BlobStats. The assigned tags are then used to remask blobs with the tag numbers (e.g., 1=TC, 2=MS, 3=SS, 4=PL, 5=others) in the StitchBlobs's output nc files.
- Finally, run TE commands demonstrated in the "additional steps" in
TE_optional.shto extract 3-hourly precipitation and 925 hPa IKE at each grid point conatined within each size/precipitation blobs that are associated with a tag number (i.e., a type of LPS).
In step four, the Python script uses a tagging arrangement like we described in the last section of the SyCLoPS manuscript. However, there are many ways one can assign those tags. In the manuscript, we define TC blobs (with tag=1) as those blobs that are paired with TC nodes in TC tracks, which corresponds to these nodes in the classified catalog:
tcid=dfc[(dfc.Short_Label=='TC') & (dfc.Track_Info.str.contains('TC'))].index.values
blobtag=np.ones(len(dfc))*5 #5 = Other systems
blobtag[tcid]=1
# Subsequent codes in the Python script: ...
However, one can also define that blobs paried with all TC nodes (not only those in TC tracks) are considered TC blobs with tag=1:
tcid=dfc[dfc.Short_Label=='TC'].index.values
blobtag=np.ones(len(dfc))*5 #5 = Other systems
blobtag[tcid]=1
# Subsequent codes in the Python script: ...
One may also define that blobs paried with all tropical LPS nodes in TC tracks are considered TC blobs with tag=1:
tcid=dfc[(dfc.Tropical_Flag==1) & (dfc.Track_Info.str.contains('TC'))].index.values
blobtag=np.ones(len(dfc))*5 #5 = Other systems
blobtag[tcid]=1
# Subsequent codes in the Python script: ...
If you are using a multiple tag system (e.g. having tag = 1,2,3,4,and more), please be careful not to have overlapping paired LPS nodes among different tags (i.e., making them all mutually exclusive). The below example shows a bad practice:
tcid=dfc[(dfc.Tropical_Flag==1) & (dfc.Track_Info.str.contains('TC'))].index.values
msid=dfc[(dfc.Tropical_Flag==1) & (dfc.Track_Info.str.contains('MS'))].index.values
blobtag=np.ones(len(dfc))*5 #5 = Other systems
blobtag[tcid]=1 #1=TCs
blobtag[msid]=2 #2=MSs
# Subsequent codes in the Python script: ...
The above codes will produce overlapped LPS nodes within tcid and msid. This will cause some TC node IDs to be overwritten by the subsequent MS IDs.
You may also just ouput one kind of tag (e.g., just tag=1) for a group of LPSs:
msid=dfc[(dfc.Short_Label.str.contains('M')) & (dfc.Short_Label=='TC') & (dfc.Track_Info.str.contains('MS'))].index.values
blobtag=np.ones(len(dfc))*0 # Other systems are all labeled 0
blobtag[msid]=1 #1=MSs
Another example:
ssid=dfc[(dfc.Short_Label=='SS(STLC)') & (dfc.Track_Info.str.contains('SS')) & ~(dfc.Track_Info.str.contains('TC'))].index.values
blobtag=np.ones(len(dfc))*0 # Other systems are all labeled 0
blobtag[ssid]=1 #1=SSs
In the above two examples, blobs that are not tagged (masked) "1" will be tagged (masked) "0." In binary masking, "0" means that blobs are not detected. Hence, the final output NetCDF blob files will only contain blobs with tag (mask)=1 associated with the desired LPS group.
After tags are assigned to blobs as described in the Python script, they will be used to alter the original blobs masks in the NetCDF files output by StitchNodes. If one groups the blob ids in terms of the tag assigned, it will look something like this:
| Tag number | Blob IDs |
|---|---|
| 1 | 50, 139, 236, 337, 438, 553, 554, 663, ... |
| 2 | 46, 137, 235, 335, 434, 436, 550, 660, ... |
| 3 | 121, 244, 709, 719, 849, 861, 935, 1153, ... |
| 4 | 1261, 1324, 1431, 1535, 1637, 1748, 1753, 185, ... |
| 5 | 1, 2, 3, 4, 5, 6, 7, 8, ... |
The output nc files with these alternations will contain blobs with their assigned tag numbers. For example, if tags 1-5 are used, grid points in each blob will be either masked 1, 2, 3, 4 or 5.
Finally, after implementing the last step (step 5) in TE, one can easily calculate the final accumulated IKE of each type of LPS over a period of time by summing each time frame of the output NetCDF files. To calculate the precipitation contribution percentage of a type of LPS, one should first calculate the total precipitation over a period by summing each time frame of the 3-hourly (or other frequency) precipitation file without any blob masks. Then do the same procedure, but with the precipitation blob masks output by TE. Lastly, the (annual/seasonal) contribution percentage of a type of LPS can be easily performed.
